evolution physiology genomics mammalogy
My research program focuses on a fundamental, unresolved issue in biology:
how genotype is related to phenotype, and the evolutionary consequences of this relationship.
Adaptive evolution often involves quantitative traits (e.g., body size, hibernation, gene expression), which are jointly shaped by many genes of small effect and the environment. I take an interdisciplinary approach to determine how relationships between genes and the environment shape complex adaptive traits. I integrate theory and techniques from evolutionary biology, physiology, molecular biology, and quantitative and population genomics to dissect how genetic architecture, organismal biology, and phenotypic plasticity interact and lead to differences in adaptive evolution.
how genotype is related to phenotype, and the evolutionary consequences of this relationship.
Adaptive evolution often involves quantitative traits (e.g., body size, hibernation, gene expression), which are jointly shaped by many genes of small effect and the environment. I take an interdisciplinary approach to determine how relationships between genes and the environment shape complex adaptive traits. I integrate theory and techniques from evolutionary biology, physiology, molecular biology, and quantitative and population genomics to dissect how genetic architecture, organismal biology, and phenotypic plasticity interact and lead to differences in adaptive evolution.
|
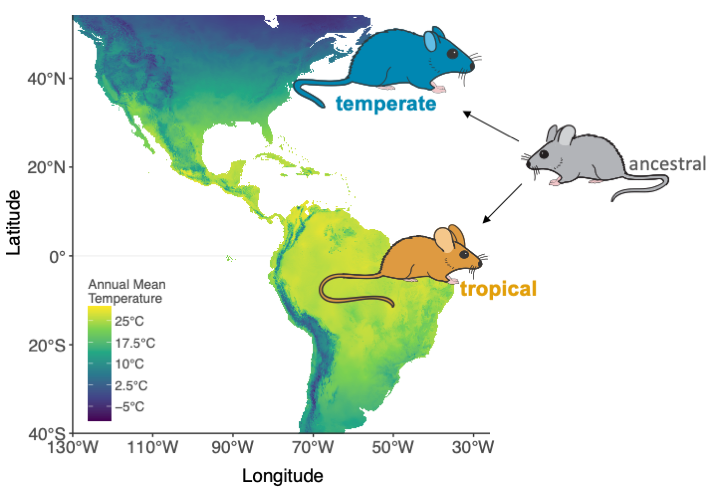
Rapid evolution to new environments Understanding how organisms adapt to their environment is a major goal in evolutionary biology. Understanding how organisms adapt to their environment is a major goal in evolutionary biology. My doctoral research investigated the genetic and environmental contributions underlying adaptation in house mice that have rapidly colonized North and South America in the last ~500 years. I discovered that house mice have quickly conformed to two of the best ecogeographic patterns in animals, as mice from tropical environments have smaller body sizes (Bergmann’s rule) and larger appendages (Allen’s rule) than mice from temperate environments (Ballinger & Nachman, 2022). Moreover, I identified the genetic basis of gene expression variation by measuring allele-specific expression (ASE; differences in expression between parental alleles) in temperate x tropical F1 hybrids across warm and cold environments. I discovered that gene expression differences between temperate and tropical house mice are largely due to cis-regulatory changes, which are robust to environmental temperature (Ballinger et al. 2022). Notably, by pairing these results with population genomic data from natural populations, I identified several cis-regulatory genes related to metabolism and body size under positive selection. Together, my results address an important knowledge gap by determining how genome function changes in response to the environment in naturally evolving, genetically diverse populations. |